VarFish is a user-friendly web application for the quality control, filtering, prioritization, analysis, and user-based annotation of DNA variant data with a focus on rare disease genetics.
It is capable of processing variant call files with single or multiple samples. The variants are automatically annotated with population frequencies, molecular impact, and presence in databases such as ClinVar. Further, it provides support for pathogenicity scores including CADD, MutationTaster, and phenotypic similarity scores. Users can filter variants based on these annotations and presumed inheritance pattern and sort the results by these scores. Variants passing the filter are listed with their annotations and many useful link-outs to genome browsers, other gene/variant data portals, and external tools for variant assessment. VarFish allows users to create their own annotations including support for variant assessment following ACMG-AMP guidelines. In close collaboration with medical practitioners, VarFish was designed for variant analysis and prioritization in diagnostic and research settings as described in the software’s extensive manual. The user interface has been optimized for supporting these protocols. Users can install VarFish on their own in-house servers where it provides additional lab notebook features for collaborative analysis and allows re-analysis of cases, e.g. after update of genotype or phenotype databases.
See our related publication in Nucleic Acids Research for details.
News
- April 13, 2021: VarFish v1.2.0 released
- First staable release of VarFish
- … also see our news entry
- April 1, 2021: VarFish v0.23.0 released
- Export ClinvarXML directly from VarFish!
- … also see our news entry
- February 12, 2021: Easy installation with Docker Compose
- get quickly started running a VarFish server using varfish-docker-compose
- … also see our news entry
- October 16, 2020: Release v0.22.0
- admins can now import arbitrary extra annotation
- adding more user interface documentation
- allow priorisation using Decipher and Orphanet disease terms
- … and dozens of more updates and fixes, see changelog for v0.22.0
Varfish Overview
VarFish provides a large set of background databases for population variant frequencies, clinical variant information, e.g., from ClinVar, gene annotations and many more. These are imported when installing VarFish. Variant calls (VCF files) are annotated and imported into VarFish. These two steps are usually performed by computationally experienced staff. Biomedical researchers can then use an easy-to-use web interface to query for and interact with the variants.
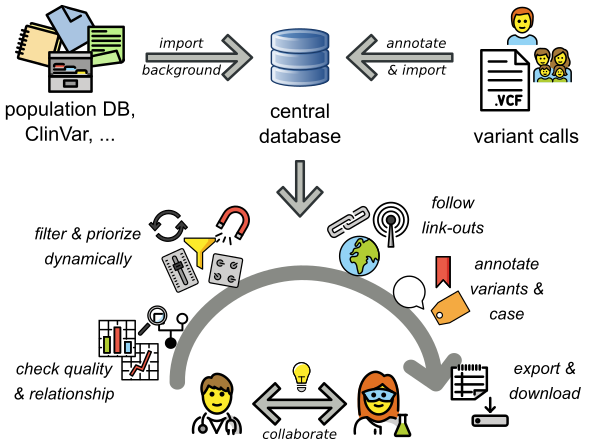
Illustration of the VarFish workflow and use cases
The software’s feature include the following:
- Various filter settings for selecting variants of interest.
- Collaborations through user commenting on and and color-flagging of variants.
- In-depth annotation of variants and genes.
- Comprehensive link-outs into public databases such as NCBI, UCSC, etc.
- Prioritization based on variant pathogenicity scores from CADD and MutationTaster.
- Phenotype-driven prioritization of genes based on HPO terms and the Exomiser software.
- Easy export and download as spreadsheet (Excel) and VCF files.
See below for our demo sites.
Software Demos
- VarFish Demo shows the features of the VarFish software including used-based commenting of variants. This is the “classic” mode where data is uploaded using a back-end API by computational staff.
- VarFish Kiosk allows users to upload their own cases as VCF files and perform an analysis.
Overall, we recommend Varfish Kiosk mostly for trying out VarFish. Many useful features such as in-house variant warehousing are turned off because of data privacy concerns and the Kiosk mode does not allow for collaboration.
Related Publications
- Holtgrewe, M.; Stolpe, O.; Nieminen, M.; Mundlos, S.; Knaus, A.; Kornak, U.; Seelow, D.; Segebrecht, L.; Spielmann, M.; Fischer-Zirnsak, B.; Boschann, F.; Scholl, U.; Ehmke, N.; Beule, D. VarFish: Comprehensive DNA Variant Analysis for Diagnostics and Research. Nucleic Acids Research 2020, gkaa241. https://doi.org/10.1093/nar/gkaa241.
Related CUBI Resources
- VarFish - VarFish instance reachable from internal networks
CUBI Contact
Last modified: Apr 12, 2022