SODAR provides
- FAIR data management for Omics studies,
- a web interface for biomedical scientists,
- command line interface and API for bioinformaticians.
For an introduction to SODAR, view the following video by Mikko Nieminen:
Features
- Scalable data storage for
- NGS raw data & metadata
- Processing Results
- Extendable metadata model, based on the ISA Tools Standard
- IGV and UCSC Integration
- Data Privacy
- Charité & MDC user authentification
- Project based authorization with different access levels
- Full audit trails
- Web-based graphical user interface for biomedical scientists
- REST API for data/systems integration
- Command line interfaces for bioinformaticians
- Data storage built on iRODS
- Metadata editor
- In use for 300+ projects with 350TB+ of stored data
Related resources and documentation
- Description of the local SODAR resource
- SODAR demo site
- SODAR manual
- BIH/MDC/Charite instance is reachable from internal networks
Technology
- Slides from the talk of Mikko Nieminen on SODAR during the 2022 iRods user group meeting
- Talk on the technology and use cases at the 2019 iRODS User Group Meeting, 30 minutes on youtube
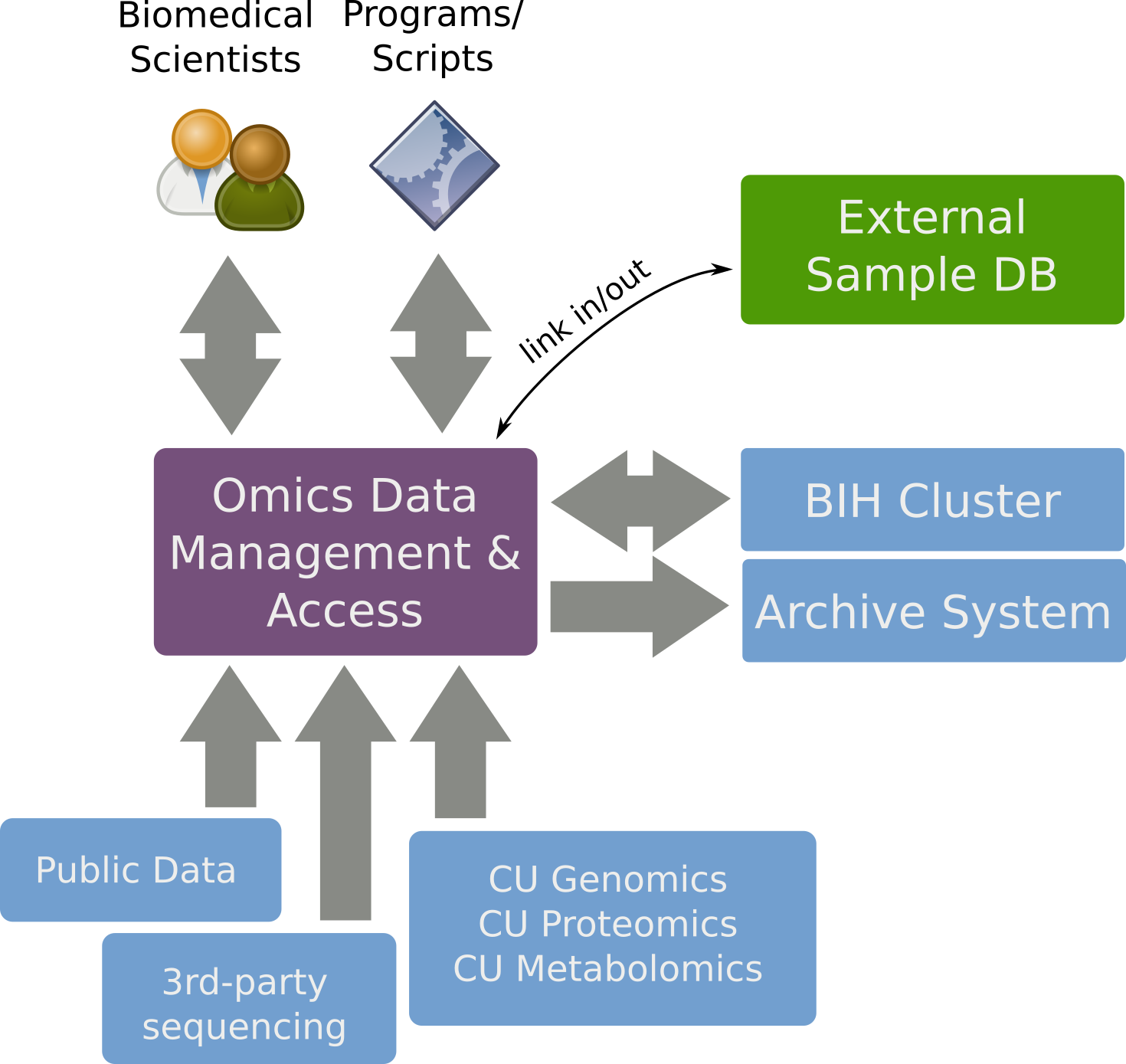
SODAR Overview
Licence and Availability
- Availability
- Published on GitHub under the MIT license
- SODAR Server repository
- SODAR Docker Compose network for evaluation, development and deployment
- Contact: Mikko Nieminen
Related Publications
Nieminen M, Stolpe O, Kuhring M, Weiner J, Pett P, Beule D, Holtgrewe M (2023). “SODAR: managing multiomics study data and metadata.” Gigascience, 12, giad052. doi:10.1093/gigascience/giad052 ☞ Link
Nieminen M, Stolpe O, Kuhring M, Weiner J, Pett P, Beule D, Holtgrewe M. SODAR: enabling, modeling, and managing multi-omics integration studies. bioRxiv. 2022 Aug 22.
M. Nieminen, O. Stolpe, F. Schumann, M. Holtgrewe, D. Beule. SODAR Core: a Django-based framework for scientific data management and analysis web apps, doi:10.21105/joss.01584
Last modified: Jul 27, 2023