A set of command line tools to call copy number variants on short read targeted sequencing panel data up to exome data. clearCNV can also re-assign samples to groups if the groups' data were generated on different panels but were confused or insufficiently documented.
Use case
Many clinical and research centers that work on reare diseases have acquired large data sets of targeted gene sequencing panel data for germline genome analysis. Such panels range from a dozen genes up to the whole exome. For such data, clearCNV allows to call copy number variants (CNVs), which are changes in the number of copies a gene is present in the sequenced genome. Such changes can be large deletions or duplications. Using the provided sequencing data, clearCNV uses this data as a background to test for clear copy number changes. Furthermore, since such data might suffer from confusion of single samples due to incomplete documentation, clearCNV is being shipped with a clustering approach to re-assign samples according to their target coverage profiles.
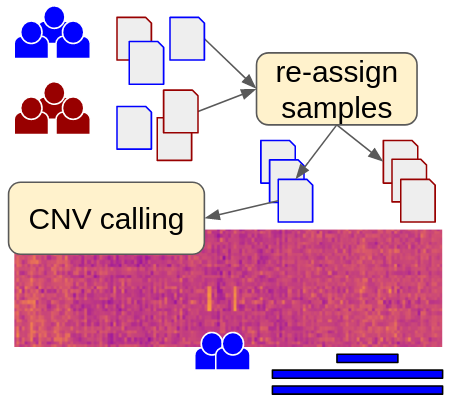
Illustration of clearCNV core use cases
Availability
CUBI Contact
Publication
- Vinzenz May and others, ClearCNV: CNV calling from NGS panel data in the presence of ambiguity and noise, Bioinformatics, Volume 38, Issue 16, August 2022, Pages 3871–3876, https://doi.org/10.1093/bioinformatics/btac418
Last modified: Aug 4, 2023