CUBI maintains and updates a growing set of resources that are designed to empower scientists and clinicians in accessing and interpreting their omics data and results.
Highlights of CUBI’s resources are shown below.
High-Performance Compute (HPC)
CUBI operates the BIH High Performance Compute (HPC) cluster containing 200+ compute nodes with ca. 6.000 cores, additional GPU and High Memory nodes and 2 PB fast parallel storage.
Further details are described on the following dedicated websites:
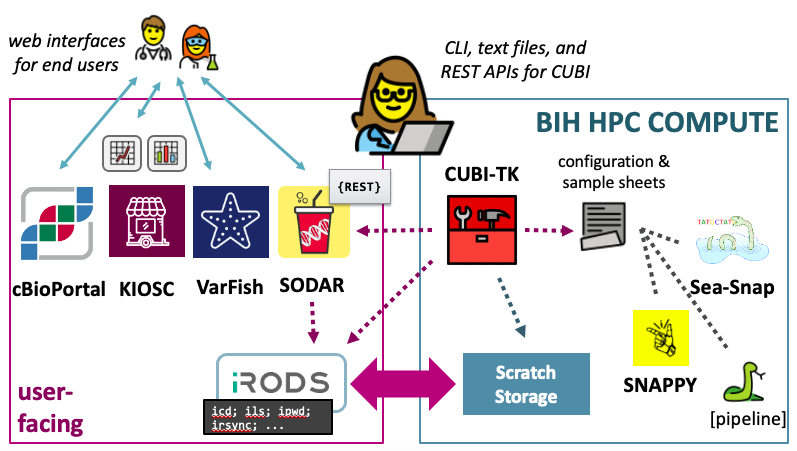
SODAR
SODAR (System for Omics Data Access and Retrieval) has been developed by CUBI to provide easy and safe access to omics data, and its metadata, processing results, and logs. SODAR is a great help in organizing data and empowering users to manage access to their data and metadata curation. The main features of SODAR include web and command line interfaces for biomedical and computational users.
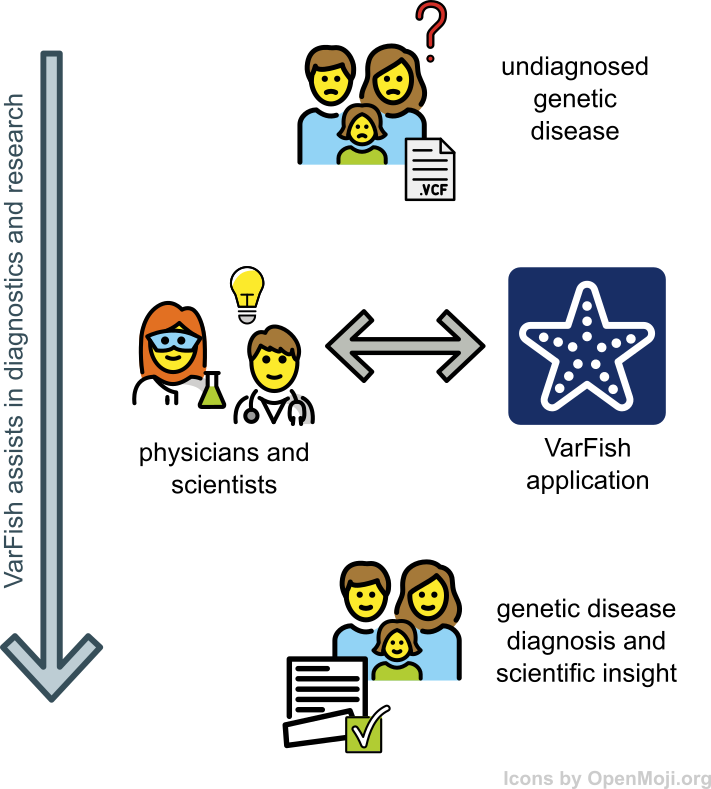
VarFish
VarFish is a data analysis platform developed at CUBI that focuses on the collaborative analysis of germline variants. We host an internal instance for use by Charite, MDC, and BIH scientists.
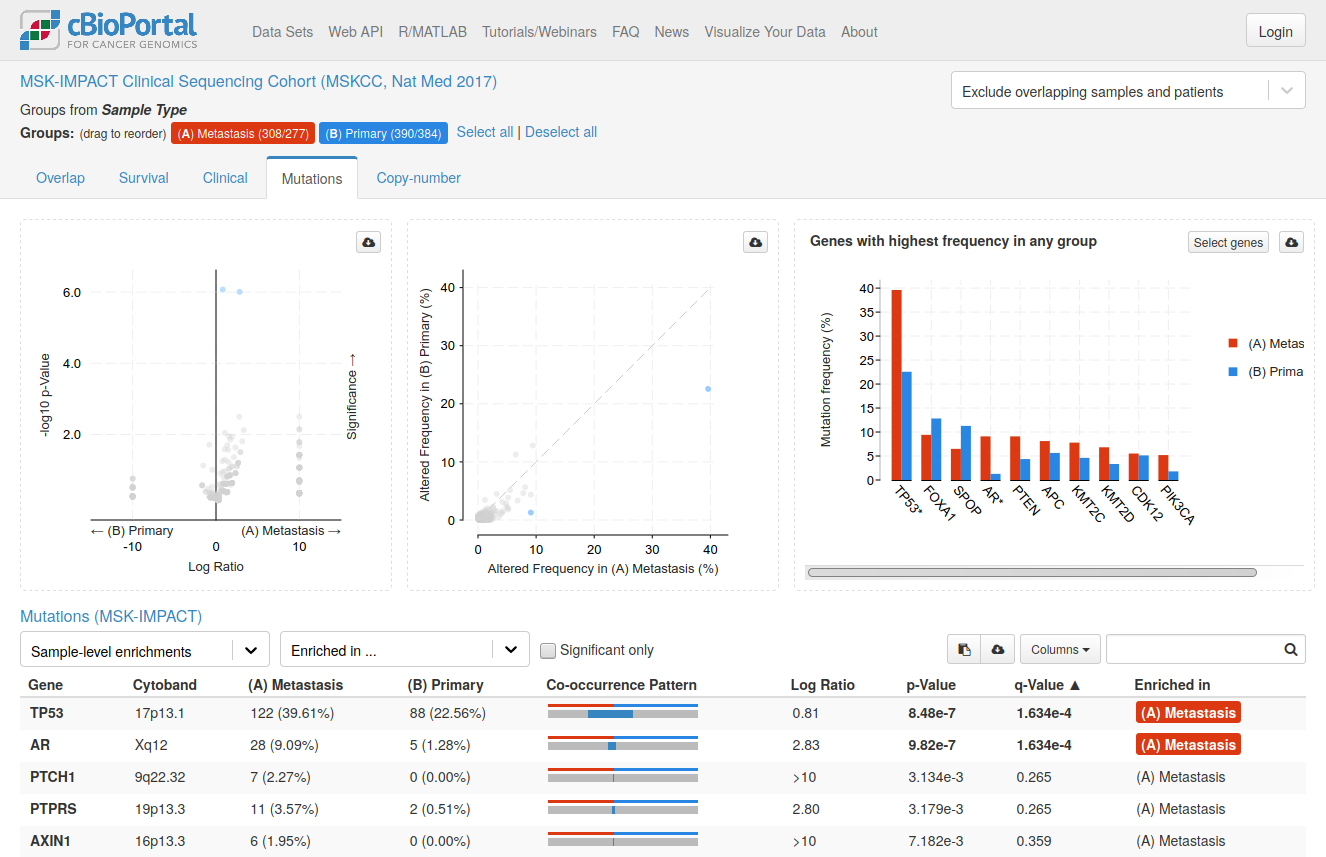
cBioPortal
cBioPortal is an interactive open-source platform that allows users to visualize and analyze genetic data from cancer studies. The platform includes the analysis of DNA mutations, gene expression, fusion genes, and copy number alterations, which can be viewed in context of clinical parameters such as sex, age, smoker/non-smoker etc.
Storage and Dataflows
CUBI is working closely together with the genomics and mass spectrometry (metabolomics and proteomics) platforms at MDC, Charite, and BIH. One area of this collaboration is managing and processing of the raw data. The initial use case is to support the genomics unit in data and meta data handling for Illumina sequencing data.